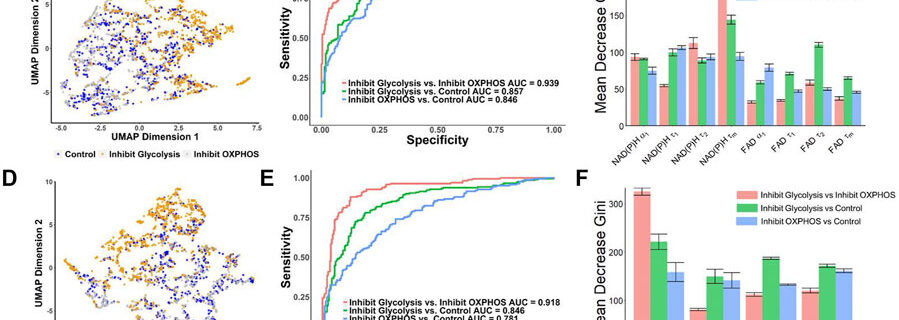
New Publication on Deep Learning to identify metabolic pathways of cells from label-free imaging
Congratulations to Walsh Lab members Linghao Hu and Daniela de Hoyos, as well as our collaborators Yuanjiu Lei and A. Phillip West (Jackson Laboratory) on our publication in APL Bioengineering, “3D convolutional neural networks predict cellular metabolic pathway use from fluorescence lifetime decay data“. In this paper, we show how label-free autofluroescence lifetime imaging of …